Get involved with our initiatives
Become involved in one of our initiatives aimed at furthering the field of top-down MS.
Please contact us to see how you can participate in our studies and projects.
Accelerating the comprehensive analysis of all human proteoforms Speeding developments in diagnostics, therapeutics, environment, and energy

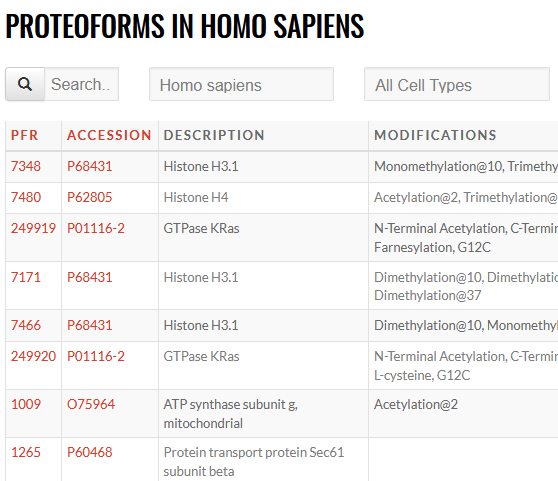
An ambitious initiative to generate a definitive reference set of the proteoforms produced from the genome, providing the bridge from genotype to phenotype

A non-profit organization with members from academic institutions, corporations, and government agencies.
With the Mission to promote collaboration, education, and innovative research in the fields of top-down mass spectrometry
Become involved in one of our initiatives aimed at furthering the field of top-down MS.
Please contact us to see how you can participate in our studies and projects.